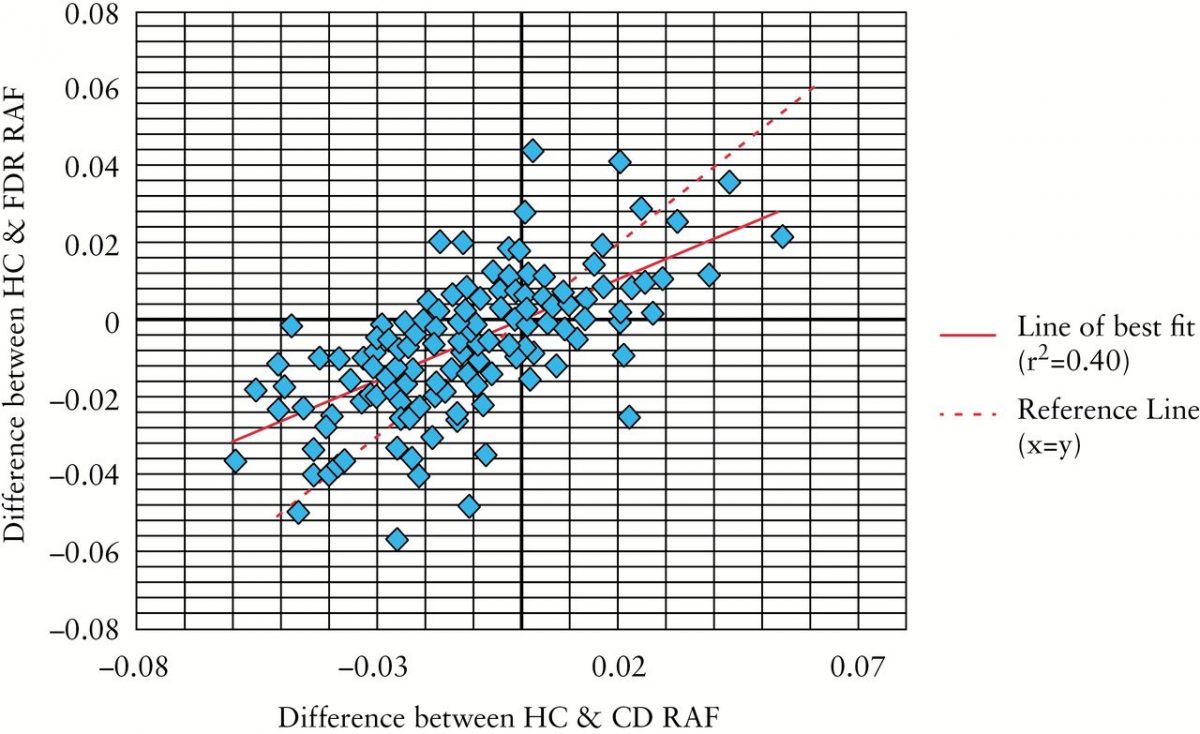
This study compared the genetic composition of healthy individuals, those diagnosed with CD, and first-degree relatives of those with CD from the GEM Project. The results of this study show that first-degree relatives of those with CD have a greater genetic risk of CD in comparison to healthy individuals.
Kevans D, Silverberg MS, Borowski K, Griffiths A, Xu W, Onay V, Paterson AD, Knight J, Croitoru K; GEM Project.
J Crohns Colitis. 2016 Feb;10(2):209-15. doi: 10.1093/ecco-jcc/jjv197. Epub 2015 Oct 28.
http://www.ncbi.nlm.nih.gov/pubmed/26512135
Abstract
BACKGROUND:
Family history provides important information on risk of developing inflammatory bowel disease [IBD], and genetic profiling of first-degree relatives [FDR] of Crohn's disease [CD]- affected individuals might provide additional information. We aimed to delineate the genetic contribution to the increased IBD susceptibility observed in FDR.
METHODS:
N = 976 Caucasian, healthy, non-related FDR; n = 4997 independent CD; and n = 5000 healthy controls [HC]; were studied. Genotyping for 158 IBD-associated single nucleotide polymorphisms [SNPs] was performed using the Illumina Immunochip. Risk allele frequency [RAF] differences between FDR and HC cohorts were correlated with those between CD and HC cohorts. CD and IBD genetic risk scores [GRS] were calculated and compared between HC, FDR, and CD cohorts.
RESULTS:
IBD-associated SNP RAF differences in FDR and HC cohorts were strongly correlated with those in CD and HC cohorts, correlation coefficient 0.63 (95% confidence interval [CI] 0.53 - 0.72), p = 9.90 x 10(-19). There was a significant increase in CD-GRS [mean] comparing HC, FDR, and CD cohorts: 0.0244, 0.0250, and 0.0257 respectively [p < 1.00 x 10(-7) for each comparison]. There was no significant difference in the IBD-GRS between HC and FDR cohorts [p = 0.81]; however, IBD-GRS was significantly higher in CD compared with FDR and HC cohorts [p < 1.00 x 10(-10) for each comparison].
CONCLUSION:
FDR of CD-affected individuals are enriched with IBD risk alleles compared with HC. Cumulative CD-specific genetic risk is increased in FDR compared with HC. Prospective studies are required to determine if genotyping would facilitate better risk stratification of FDR.
Copyright © 2015 European Crohn’s and Colitis Organisation (ECCO). Published by Oxford University Press. All rights reserved. For permissions, please email:
[email protected].